Chloe Hsu
I completed my PhD at the University of California, Berkeley, advised by Jennifer Listgarten and Moritz Hardt. |
Research |
|
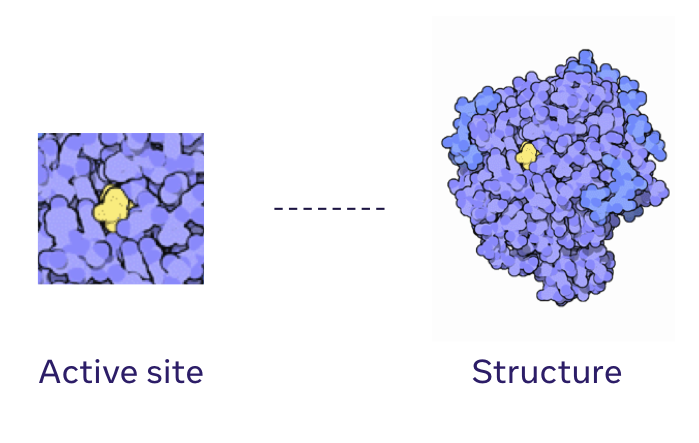
Generative models for protein structures and sequencesChloe Hsu, Clara Fannjiang, and Jennifer Listgarten Nature Biotechnology, 2024 paper How can generative models be useful for protein engineering? |
|
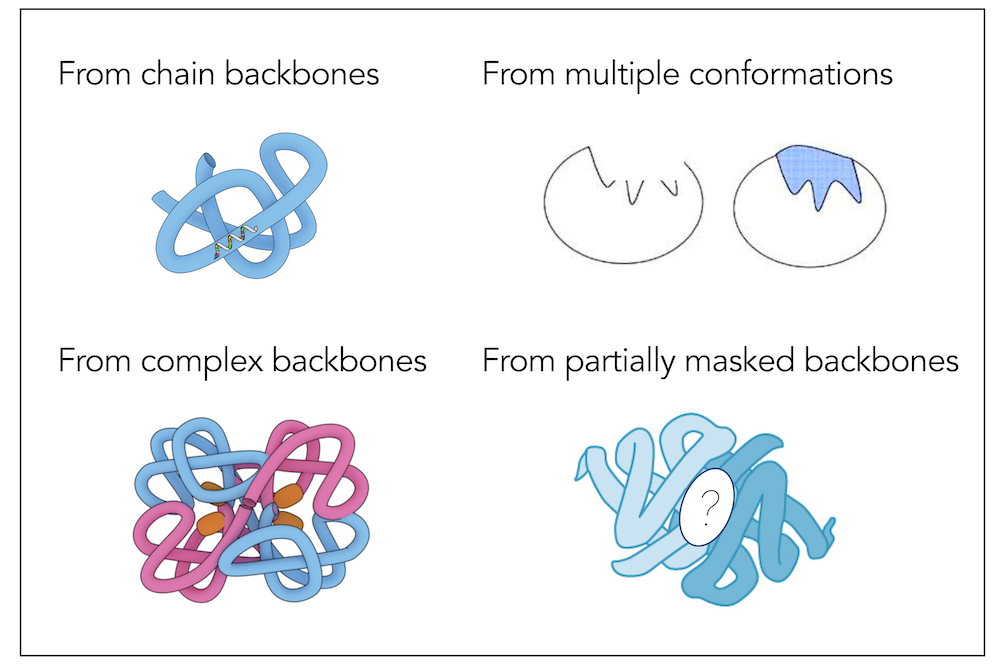
Learning inverse folding from millions of predicted structuresChloe Hsu, Robert Verkuil, Jason Liu, Zeming Lin, Brian Hie, Tom Sercu, Adam Lerer*, Alexander Rives* ICML (Outstanding Paper Runner Up Award), 2022 paper | code | slides | colab notebook Inverse folding aims to design sequences to fold into desired structures. With 12M new predicted structures as additional training data, ESM-IF1 is more accurate at structure-based sequence design, while also generalizing to more sophisticated design tasks. |
|
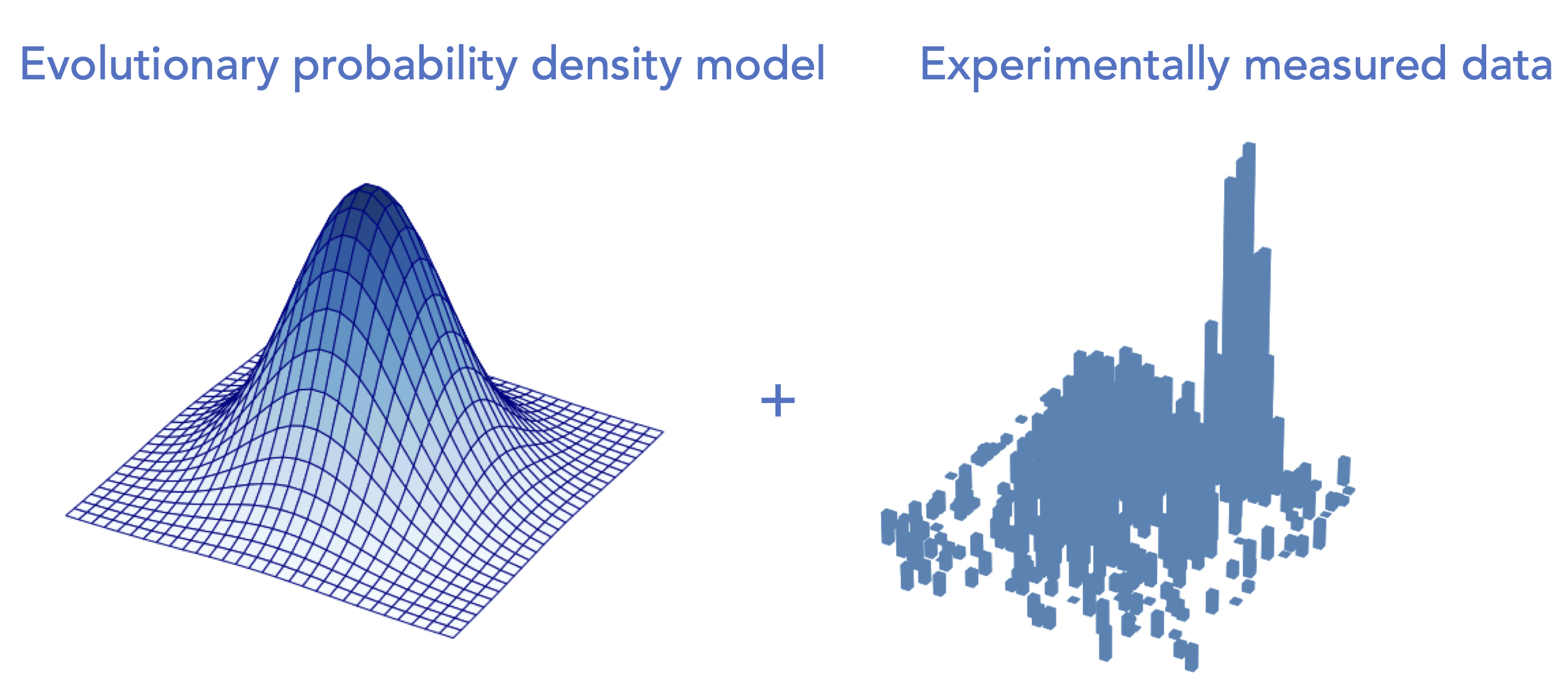
Learning protein fitness models from evolutionary and assay-labeled dataChloe Hsu, Hunter Nisonoff, Clara Fannjiang, and Jennifer Listgarten Nature Biotechnology, 2022 paper | talk | code A simple yet highly effective hybrid approach to protein fitness prediction. Also a comparative analysis to highlight the importance of systematic evaluations and sufficient baselines. |
|
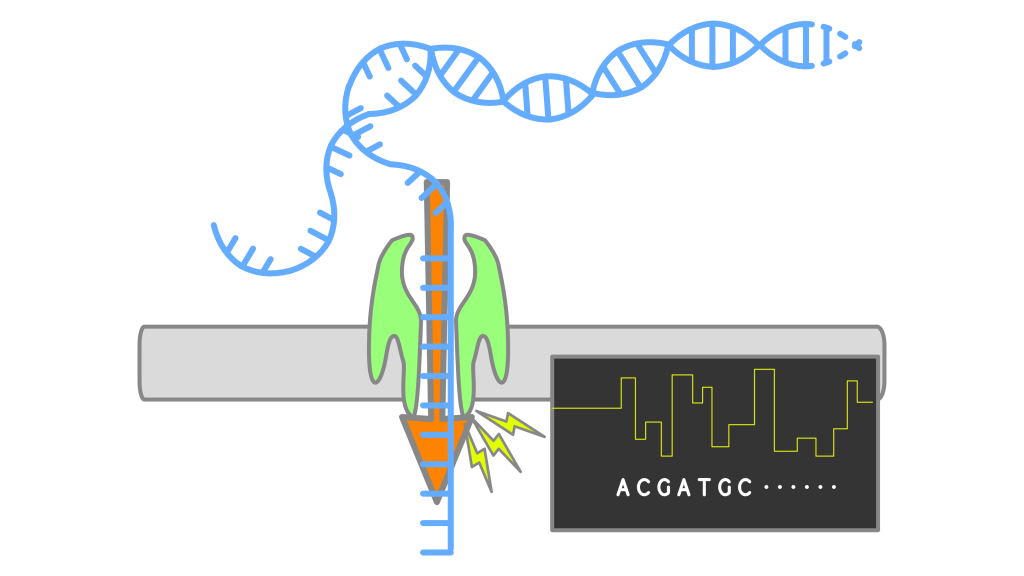
Nanopore callers for epigenetics from limited supervised dataBrian Yao, Chloe Hsu, Gal Goldner, Yael Michaeli, Yuval Ebenstein, and Jennifer Listgarten bioRxiv, 2021 paper Calling epigenetic modifications on nanopore sequencing platforms when the training data is incomplete. |
|
Design and source code from Jon Barron's website and Leonid Keselman's Jekyll fork. |